Difference between Restriction Endonuclease and Exonuclease
Restriction Endonucleases, also known as restriction enzymes, cut DNA from the inside. These enzymes are found in bacteria to protect against viruses. Exonucleases, on the other hand, cut the DNA from the ends. It is an important topic in biology.
This Story also Contains
- What is Restriction Endonuclease and Restriction Exonuclease?
- What are Nucleases?
- Difference between Endonuclease and Exonuclease
- Restriction Endonuclease Enzyme
- Example of Restriction Endonuclease
- Application of Restriction Endonuclease
- Restriction Exonuclease Enzyme
- Function of Restriction Exonuclease
- Practical Applications of Restriction Endonucleases in Molecular Biology
- Recommended Video:
The main function of these enzymes is to divide the DNA at specific points. The functions and types of different restriction enzymes, along with their examples, are really important for the preparation of NEET, AIIMS, Nursing, and paramedical exams. This article covers the details about restriction endonuclease, restriction exonucleases, their differences, and different types of restriction endonucleases.
What is Restriction Endonuclease and Restriction Exonuclease?
Restriction endonucleases divide the DNA molecules internally at a specific site, whereas exonucleases remove nucleotides from the ends of DNA or RNA molecules and are involved in genetic engineering and molecular biology. Nucleases are therefore important in the manipulation of nucleic acids because they link the phosphodiester bonds within DNA and RNA.
Due to the many roles of nucleases in genetic engineering and molecular biology, it is important to learn about restriction endonucleases, exonucleases, and other nucleases. Among them, restriction endonucleases, which cut DNA at specific sites, are necessary for cloning, gene engineering, and recombinant DNA technology, whereas exonucleases, which remove nucleotides from the ends of DNA, are useful for DNA repair as well as replication.
The enzymes that can rather accurately and selectively divide, splice, or join nucleic acids have thoroughly changed molecular biology and encouraged the creation of techniques like CRISPR and gene therapies; the latter may be evidence of how these enzymes are valuable in research and industry.
Also Read:
What are Nucleases?
Nucleases are enzymes that hydrolyze the phosphodiester bond of nucleic acids, leading to DNA cleavage into small pieces. The enzyme mechanism involves DNA and RNA metabolism, such as DNA repair, replication, and recombination, as well as RNA decay. Nucleases can be further categorised based on their results on the nucleic acid chain, with examples being endonucleases that cut at a particular site on the chain and exonucleases that break down the DNA from the ends.
Difference between Endonuclease and Exonuclease
There are certain differences between the endonuclease and the exonuclease, which separate them from one another. Let us differentiate between restriction endonucleases and exonucleases based on the features in the table below.
Feature | Restriction Endonucleases | Exonucleases |
Definition | Enzymes that cut DNA at specific recognition sites | Enzymes that remove nucleotides from DNA end |
Site of Action | Internal DNA sequences | DNA ends (5’ or 3’) |
Mechanism | Recognise and cleave specific DNA sequences | Trim nucleotides sequentially from DNA ends. |
Product obtained | The product obtained after cleavage is an oligonucleotide chain. | The product obtained after exonuclease activity is a monomer of nucleotides. |
Types | Type I, II, III, and IV | 5' to 3' exonucleases, 3' to 5' exonucleases |
Applications | Cloning vector, gene editing, and recombinant DNA | DNA repair, removal of RNA primers, DNA replication |
Lag Period | It shows a lag period during their activity. | It does not show any lag period. |
Example | EcoRI, HindIII | Exonuclease I, Exonuclease III |
Recognition Sequence | Specific palindromic sequences | Not sequence-specific |
Cutting Pattern | Specific cuts produce sticky or blunt ends | Sequential removal of nucleotides |
Usage in Laboratories | Creating recombinant DNA, restriction mapping | DNA repair studies, degradation of RNA primers |
Restriction Endonuclease Enzyme
Restriction endonuclease, also called restriction enzymes, involve enzyme mechanisms that hydrolyse DNA at specific restriction sites that exist within a DNA molecule. They are widely used in genetic engineering because they cut the DNA at specific places for insertion or alteration of genes.
Restriction endonucleases were for the first time identified in the early 1960s during research on the strategies used by bacteria to fight against viral infection. Hamilton Smith and his co-workers named the first restriction enzyme discovered as HindII, and it existed in the bacterium Haemophilus influenzae. This finding transformed molecular biology and was the foundational innovation for constructing genetically tailored modern technology.
Types of Restriction Endonuclease (Type I, II, III, and IV)
There are four types of restriction endonucleases:
Type I: These enzymes only bind to some DNA sequences, but cut at any position that is arbitrary relative to the binding site.
Type II: These enzymes can bind to the DNA sequence specific to them and cut at particular locations within or just next to the recognition sequence. REases are classified into four groups; Type I, II, III and IV, but only type II REases are widely used in molecular biology.
Type III: These enzymes can bind with the DNA sequence, and the DNA gets cut at a fixed distance from the binding site.
Type IV: These enzymes cut DNA at particular sites, but for this procedure, they do not need ATP hydrolysis.
Function of Restriction Endonucleases
The mechanism of action and function of restriction endonuclease are given below:
Recognition sites
Restriction endonucleases come into contact with the target DNAs by the direct complementary base interactions with the preferred, usually palindromic, nucleotide sequences. After becoming bound, the enzyme causes the breakdown of phosphodiester bonds within the DNA structure, thus causing DNA cleavage and dividing it into two sections.
Cleavage patterns
The specific sequences recognized by restriction endonucleases are normally palindromic, which simply means that they read the same, from the 5’ to 3’ direction and in a similar sequence from the 3’ to the 5’ direction. For instance, the specific recognition site of restriction enzyme EcoRI is 5’ GAATTC 3’, it is the same for both strands of the DNA helix if read from 5 ’to 3’. The cutting patterns of restriction endonucleases differ with the enzyme under consideration as well as the particular recognition sequence. Some enzymes cut the DNA through both chains in a straight cut, while others make an angular cut that gives rise to overhanging or ‘sticky’ ends.
Example of Restriction Endonuclease
One of the more commonly known restriction endonucleases is EcoRI, the very first restriction enzyme isolated. This cuts DNA at a specific recognition sequence - GAATTC - that allows it to be an important tool for molecular biology in dealing with DNA manipulation.
Application of Restriction Endonuclease
There are a few really important applications of restriction endonucleases. From genetic engineering and cloning to DNA mapping, it works as a savior. The application of Restriction endonuclease enzymes is given below:
Genetic engineering
Restriction endonucleases are used to divide the DNA at definite sites so that the foreign DNA fragments can be inserted or the unwanted sequences can be deleted.
Cloning
They are also involved in the formation of recombinant DNA molecules through their ability to make V-shaped cuts at the restriction recognition site of the vector and insert DNA.
DNA mapping
In molecular biology, methods like restriction fragment length polymorphism analysis (RFLP) and Southern blotting, restriction endonuclease enzymes are used to determine the sequence of DNA strands and variations, respectively.
Restriction Exonuclease Enzyme
Exonucleases are enzymes that remove nucleotides through endonucleases provided in the nucleic acids of both DNA and RNA. They have important functions related to DNA repair, replication, and RNA processing because they generate and recognise abasic sites or any nucleotide lesion.
Restriction Exonuclease Diagram
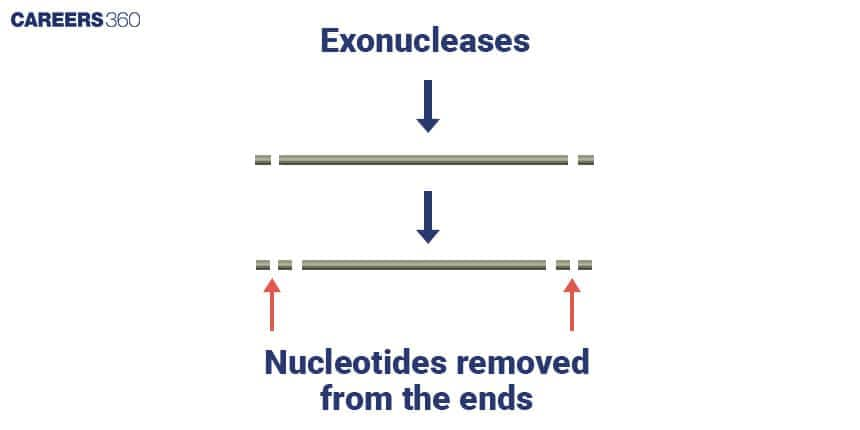
Types of Restriction Exonuclease
There are two main types of exonucleases:
5’ to 3’ exonucleases
These enzymes remove nucleotides, one at a time, from the 5’ end of the DNA or RNA strand.
3’ to 5’ exonucleases
These enzymes cut nucleotides off the 3’ end of the DNA or RNA strand in a stepwise manner.
Function of Restriction Exonuclease
The enzyme mechanism involves exonucleases participating in the breakdown of a phosphodiester bond of the nucleotide. This results in the release of nucleotides at the terminal of the DNA or RNA strand. They also have polarity depending on which direction of the nucleic acid strand they originate from.
Example of Restriction Exonuclease
One of the common restriction exonucleases is ExoI. This enzyme degrades single-strand DNA from the 3' end and is implicated in DNA repair and recombination processes.
Application of Restriction Exonuclease in Molecular Biology
Exonucleases have multiple applications in molecular biology:
DNA repair
Exonucleases participate in the repair processes of DNA, which include base excision, and nucleotide excision processes for the removal of damaged, mismatched nucleotides in the DNA strands.
Degradation of RNA primers
Within the DNA replication process, RNA primers are utilised and eventually erased by exonucleases so that DNA polymerase can extend the newly created strand with DNA nucleotide.
Removal of mismatched nucleotides
Exonucleases interact with DNA mismatch repair pathways to help remove nucleotides that fail to pair with the template DNA strand in the right way thereby maintaining genome stability.
Practical Applications of Restriction Endonucleases in Molecular Biology
Restriction enzymes laid the foundation for precise gene editing, which gave the way for advanced technologies like CRISPR-Cas9. A few important applications of restriction endonucleases are given below:
Genetic Engineering Techniques Involving Restriction Endonucleases
Restriction endonucleases are necessary in the process of genetic manipulation, as they allow for the fine control of DNA fragments. Processes like Recombinant DNA technology use restriction enzymes to cut DNA at designed positions with a view to meaningful insertions of foreign DNA segments into vector molecules. Restriction endonucleases are used in all the processes of gene cloning, gene editing, and gene expression to produce genetically modified organisms and to study the function of genes.
DNA sequencing and mapping
Restriction endonucleases are commonly used in DNA sequencing and mapping activities. Molecular techniques. This includes RFLP analysis and Southern blotting, which involve the use of restriction enzymes that cut the DNA at specific sites, producing typical patterns of fragments that can be used to establish a relationship between genes and chromosomes in addition to revealing genetic variations and thus studying the DNA structure and organisation.
Role Of Exonucleases in DNA Replication and Repair
Genomic exonucleases have important functions in DNA replication and repair. They help in protecting the integrity of the genome. During DNA replication, RNA primers are cleaved from the newly synthesised DNA chains by exonucleases and DNA polymerases, which then use DNA nucleotides to fill the gap. In DNA repair, exonucleases eliminate several nucleotides in a single type, mismatched to other nucleotides in a strand of DNA, by participating in the process of repairing errors and maintaining the integrity of genes.
Case Study: Use of Restriction Enzymes in CRISPR Technology
CRISPR, or Clustered Regularly Interspaced Short Palindromic Repeats, is a function that uses restriction enzymes for editing genes. CRISPR-associated (Cas) proteins are a group of proteins that function as nucleases that are programmable, in which they are guided by short RNA sequences to the target DNA.
With the help of a guide RNA (gRNA) that complements the targeted DNA sequence, the Cas nuclease cuts the DNA at the specific site. This tool has helped in gene editing and has changed molecular biology because it allows scientists to work on genes with increased accuracy and speed.
Both restriction endonucleases and exonucleases are very important elements in molecular biology that are being used in areas like genetic manipulation and DNA sequencing, DNA replication, and DNA repair.
Recommended Video:
Frequently Asked Questions (FAQs)
Restriction endonucleases’ main purpose is to cut DNA at specific known sequences, thus contributing to genetic engineering by providing methods to alter DNA molecules.
Exonucleases are not the same with restriction endonucleases concerning their modes of operation. Restriction endonucleases act inside the DNA molecule, breaking the chain at certain points, while exonucleases modify the molecule by eradicating nucleotides progressively, beginning from the ends of the chain, be it DNA or RNA
Restriction endonucleases are useful in several techniques of genetic engineering, such as gene cloning, gene manipulation and DNA sequencing. They are used to cleave the DNA at certain sites to enable the addition or deletion of certain parts of the DNA.
Exonucleases are also involved in DNA repair because they will be able to eliminate nucleotides that have correct sequences or are in some way damaged on one of the DNA strands. Nonetheless, their primary role is not limited to the context of DNA repair, in contrast to other enzymes enclosed in the framework of repairing mechanisms.
Restriction endonucleases and exonucleases, however, display contrasted specificities and modes of action in terms of cleavage. Restriction endonucleases cut the DNA at a particular sequence, while restriction endonucleases degrade the DNA or the RNA from the ends, 5’ to 3’ end or 3' to 5’ end.
The first restriction endonuclease isolated was HindII, which was discovered in 1970. It cuts DNA at specific recognition sites, making it a pivotal discovery in the field of molecular biology.