Transcription in Prokaryotes: Process, Steps, Diagram
Definition Of Transcription
Transcription itself is the biological process where, using the enzyme RNA polymerase, the information from DNA gets replicated into messenger RNA. This is an archetypal component of the translation of genetic instructions into functional proteins.
In prokaryotes, gene expression and regulation are based on transcription. This process allows prokaryotic microorganisms to respond as soon as possible—or at another level—to differentiation in their exterior environment through certain proteins that are necessary for their survival and adaptation.
Description Of Transcription Process
NEET 2025: Mock Test Series | Syllabus | High Scoring Topics | PYQs
NEET Important PYQ's Subject wise: Physics | Chemistry | Biology
New: Meet Careers360 B.Tech/NEET Experts in your City | Book your Seat now
Prokaryotic transcription is described overall by initiation, elongation, and termination. The steps are realised with the help of specific proteins and DNA sequences to ensure accurate and efficient transcription.
Prokaryotic Cell Structure
Prokaryotic cells are single cells that lack a membrane-bound nucleus. The location of the genetic material makes these relatively very simple in structure compared to those of eukaryotic cells.
Components Involved In Transcription
DNA: The template for the synthesis of RNA. In prokaryotes, DNA is usually a single, circular chromosome.
RNA Polymerase: This enzyme is responsible for synthesizing RNA from the DNA template.
Promoter Regions: These are specific DNA sequences that mark the beginning of a gene and specify the binding site for RNA polymerase.
Transcription Process In Prokaryotes
The steps of the transcription process in prokaryotes are:
Initiation
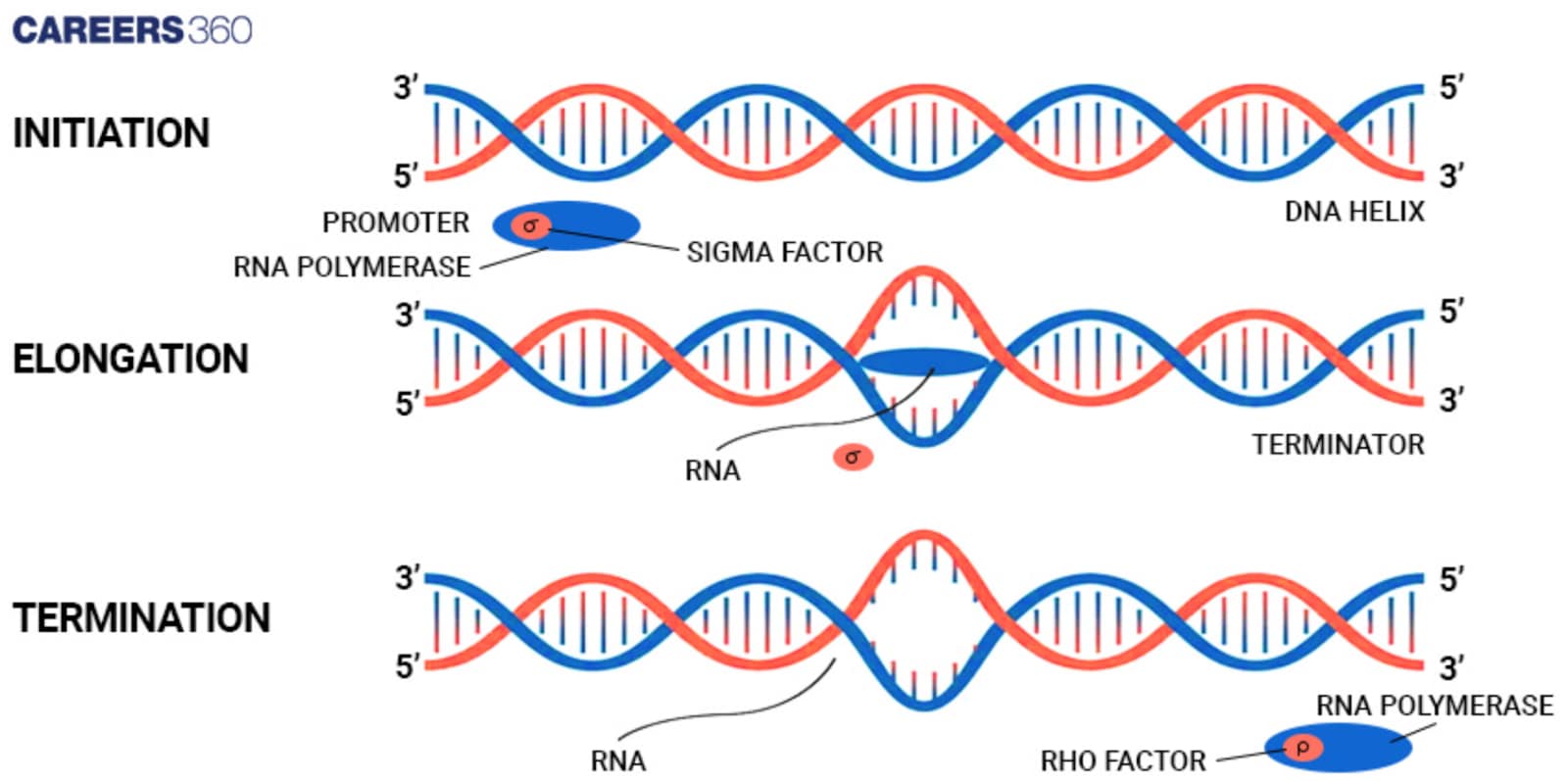
Role of Sigma Factor: This would be the subunit of RNA polymerase to recognize and bind promoter regions on DNA. It would ensure that RNA polymerase attaches to an appropriate start site for transcription.
Binding to the Promoter Region: The RNA polymerase-sigma factor complex binds to the promoter region. Binding itself induces local unwinding of the DNA to form the so-called open complex. This unwound segment of DNA is where the initiation of transcription will occur.
Elongation
The Transcription Bubble Formation: While RNA polymerase travels along the DNA, it unwinds the double helix continuously, making available the template strand and forming a 'transcription bubble'.
Synthesis of mRNA: The RNA polymerase reads the template strand of the DNA and synthesizes a complementary strand of RNA by the addition of RNA nucleotides. The strand of RNA elongates in a direction from 5' to the 3'
Termination
Rho-dependent Termination: Rho protein will bind to the RNA and translocate on it toward the RNA polymerase. When it catches up with the polymerase, it causes the release of the enzyme from the RNA, thereby terminating transcription.
Rho-independent Termination: This depends on specific sequences in the RNA, which form a hairpin structure followed by a series of uracil nucleotides. This structure causes RNA polymerase to pause and fall off the DNA thus terminating transcription.
Diagram: Transcription In Prokaryotes
The diagram given below shows the process of transcription
Regulation Mechanisms Of Prokaryotic Transcription
Operons And Their Role
Lac Operon: It is the most straightforward operon system in control of the transcription of genes involved in lactose metabolism. The system contains a promoter, operator, and structural genes. Control of the lac operon is regulated by both positive and negative control mechanisms.
Positive And Negative Regulation
Positive control refers to the activator proteins that facilitate the binding of RNA polymerase to the promoter thereby enhancing the rate of transcription.
Negative Repressor proteins bind to the operator region and do not allow the RNA polymerase to start transcription.
Attenuation Mechanism
A kind of regulatory mechanism under which the transcription is terminated prematurely depending on the secondary structure formation in mRNA. This helps in fine-tuning the control of expression according to the environmental conditions in further detail.
Conclusion
Transcription in prokaryotes takes place in three stages: initiation, elongation, and termination of RNA synthesis. The key components behind this complex process are RNA polymerase, the sigma factors, promoter regions, and various regulatory proteins. The process is highly regulated to bring about accurate expression of genes.
Understanding transcription in the prokaryotes is important for clarifying the fundamental principles of biological processes, bacterial adaptation, and the control of genes. It also has big implications for biotechnology and medicine in the development of antibiotics and genetic engineering.
Future directions of research may lie within the detailed mechanisms of transcription regulation, researching transcription in various prokaryotic species, and exploring its potential for manipulation in biotechnological applications. With new methodologies being developed, new knowledge of high-resolution imaging and genome editing tools will advance this research.
Recommended video for "Prokaryotic Transcription"
Frequently Asked Questions (FAQs)
Transcription is the process whereby a portion of DNA gets duplicated into RNA through RNA polymerase.
In prokaryotes, the process of transcription is simple and only involves one type of RNA polymerase, while in eukaryotes, it is those that are complex, involving three kinds of RNA polymerases with some extra processing steps.
The sigma factor guides RNA polymerase to recognise and bind the DNA promoter regions and initiates the process of transcription.
Rho-dependent termination and Rho-independent are two of the main types of termination in prokaryotes.
The lac operon is subject to regulation by negative and positive control mechanisms, through the lac repressor protein and the availability of lactose.
Also Read
29 Nov'24 09:31 AM
19 Nov'24 09:26 AM
18 Nov'24 06:45 PM
18 Nov'24 09:29 AM
18 Nov'24 09:18 AM
18 Nov'24 09:01 AM
18 Nov'24 08:37 AM
16 Nov'24 03:45 PM