DdNTP Full Form

What is the full form of DdNTP?
DdNTP, or Deoxyribonucleotide Triphosphate, are artificially prepared nucleotides. They are chemical compounds having a pentose sugar, a nitrogenous base, and three phosphate molecules. The pentose sugar here is deoxyribose, i.e., ribose sugar deoxygenated twice. Due to the absence of hydroxyl groups at both the 2' and 3' locations on the ribose, they are also known as 2' and 3'. They can be of multiple types, depending on the type of nitrogenous base attached. The three phosphate molecules are attached by high-energy bonds. Their main use is in Sanger sequencing for the purpose of chain terminalization. They help prevent chain elongation, thus keeping the number of nucleotides in a DNA fragment under control.
Nucleotide
A nucleotide is the smallest unit, or the building block, of DNA. It is composed of primarily three units- a pentose sugar, a nitrogenous base and phosphate molecule(s). They are of two types on the basis of type of sugar base- RNA (which contains ribose sugar) or DNA (which contains deoxyribose sugar). The nitrogenous bases are attached to the second member of pentose sugar using a glycosidic bond, and the phosphate group is attached to the fifth carbon of sugar by a phosphodiester bond. Two nucleotides are joined by phosphodiester bonds.
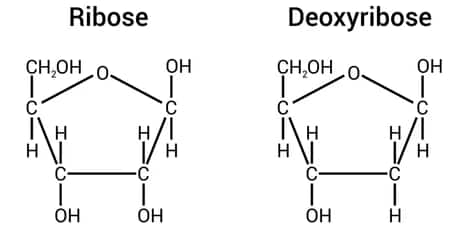
Figure: Ribose and Deoxyribose sugar
Sugar Base
The nucleotide is made up of a pentose sugar. This may be ribose sugar or deoxyribose sugar. A pentose sugar is a 5-carbon compound with a hydrogen and a hydroxyl attached to carbon atoms, which are then bonded to adjacent carbons. Two carbon atoms are attached to an oxygen atom in the ring structure. The deoxyribose and ribose sugars differ in the sense that the former lacks a hydroxyl group on carbon number 2. The deoxyribose sugar, however, lacks a hydroxyl group on two carbon atoms as compared to the sugar, which has carbons 2 and 3.
Nitrogenous Base
Biological molecules containing nitrogen are known as nucleobases, often known as nitrogenous bases or frequently just bases. There are two categories of nitrogenous bases: purines and pyrimidines. Under purines, there are two types of nitrogenous bases: adenine and guanine, while under pyrimidine, there are three types: cytosine, thymine, and uracil. The nitrogenous bases are attached to the first carbon of the pentose sugar by a glycosidic bond. The names of the respective nitrogenous bases and their nucleotides are as given below:
Nitrogenous base | Nucleotide |
Adenine | Adenosine |
Guanine | Guanosine |
Cytosine | Cytidine |
Thymine | Thymidine |
Uracil | Uridine |
Chain Polymerisation
Nucleotides are joined by phosphodiester bonds in order to form a DNA strand. A phosphate group is attached to the fifth carbon of each sugar base. This phosphate group bonds with the hydroxyl group present on the third carbon of the next pentose sugar. The bond is formed between these two groups while a molecule of water is released. This is the phosphodiester bond. Therefore, for the formation of a chained polymer of DNA or RNA, the presence of at least one phosphate group on fifth Carbon of sugar and that of hydroxyl group on third Carbon is required.
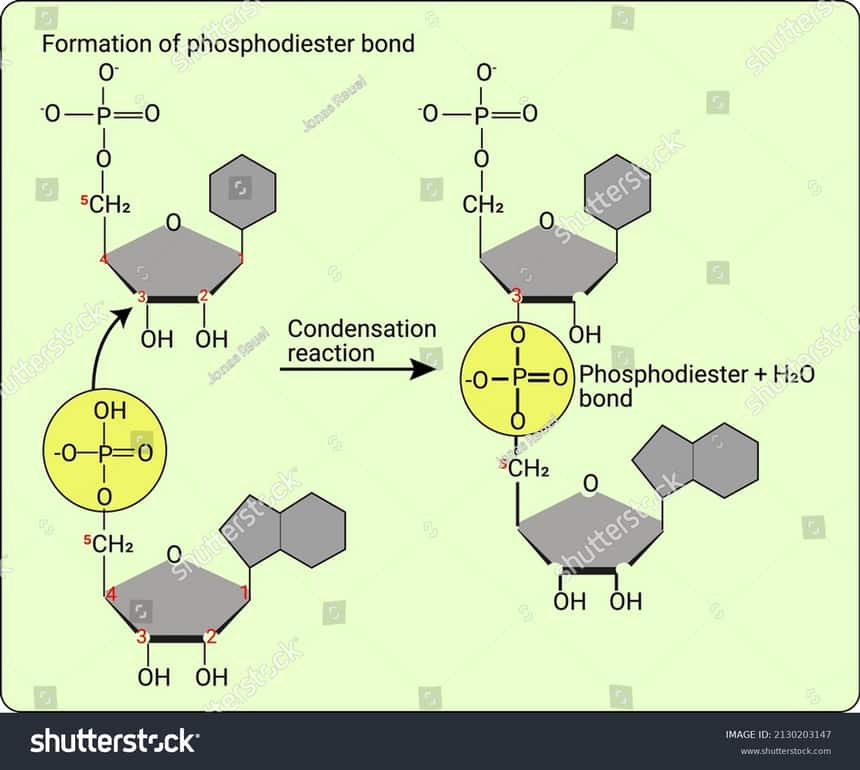
Figure: Formation of phosphodiester bond
Sanger sequencing
Developed by Frederick Sanger, it is a method of determining the sequence of nucleotides in DNA fragments.
The following are the steps involved in Sanger DNA sequencing:
(1) Denaturation of the double-stranded DNA (dsDNA) results in the formation of two single-stranded DNAs (ssDNA).
(2) Attached to the sequence is a primer that matches one end.
(3) Only one type of ddNTP and four different types of dNTPs are added to each of the four polymerase solutions.
(4) The DNA synthesis reaction starts, and the chain grows until an unpredicted termination nucleotide is added.
(5) Denatured ssDNA is created from the resultant DNA pieces.
(6) Gel electrophoresis is used to separate the denatured fragments and identify their sequencing.
Chain terminalization
Sanger sequencing requires both deoxyribonucleoside triphosphates (dNTPs) and their corresponding derivatives, dideoxyribonucleoside triphosphates (ddNTPs). During polymerization, dNTPs function as monomers, while ddNTPs are employed for chain terminalization. Since phosphodiester bonds cannot be formed with the next nucleotide, the 3' H present in ddNTPs inhibits chain elongation.
ddNTP is helpful in analysing the structure of DNA because it prevents a DNA strand from polymerizing during DNA replication, resulting in varied lengths of DNA strands duplicated from a template strand. Later, these freshly created DNA strands are employed in gel electrophoresis to create a series of band patterns that are helpful to determine the DNA strand's sequence.
Types of ddNTP
On the basis of the type of nitrogenous base attached to the first carbon of the pentose sugar, ddNTPs can be segregated into the following types:
Nitrogenous Base | ddNTP |
Adenine | dideoxyadenosine-5′-triphosphate |
Guanine | deoxyguanosine-5′-triphosphate |
Cytosine | deoxycytidine-5′-triphosphate |
Thymine | dideoxythymidine-5′-triphosphate |
Difference between dNTP and ddNTP
Basis of differentiation | dNTP | ddNTP |
Structure | Hydroxyl group on the third carbon of pentose sugar. | The hydroxyl group on the third carbon of pentose sugar is absent. |
Use | Monomer for DNA formation. | Used in chain terminalization during Sanger sequencing. |
Bonding | It is capable of forming phosphodiester bonds with adjacent nucleotides. | It is incapable of forming phosphodiester bonds with adjacent nucleotides. |
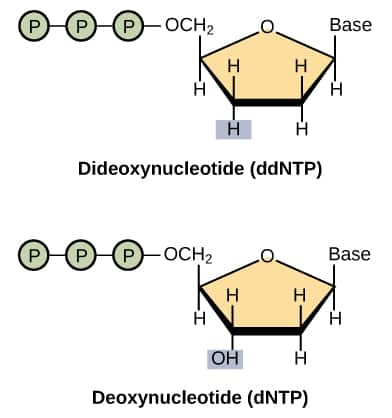
Figure: Difference in structure of dNTP and ddNTP
Frequently Asked Questions (FAQs)
ddNTP is dideoxynucleotide triphosphate, whereas dNTP is deoxynucleotide triphosphate. Both of these are thus nucleotides, i.e., they contain a pentose sugar, a phosphate group, and a nitrogenous base. Also, both of them are used in DNA synthesis.
PCR, or polymerase chain reaction, is used to detect disease in its early stages by detecting the pathogen after replicating the DNA multiple times. Since replication of DNA is involved, Sanger’s method is used. A target DNA segment is amplified using the Sanger method in order to precisely ascertain the DNA sequence. Simply put, the integration of ddNTPs in the reaction mechanism serves to stop the synthesis of an expanding DNA strand, leaving behind DNA fragments that have partially duplicated.
The distribution of DNA fragment lengths produced by Sanger sequencing depends on the ratio of dNTP to ddNTP being used.
The product will be shorter and closer to the primer if this ratio increases, whereas the product will be longer and farther from the primer if it decreases.
Gel electrophoresis is used to separate these DNA fragments, and the length of each fragment may be measured in relation to how far it has travelled through the gel using autoradiography. The band that travelled the farthest through the gel must have been the sequence that terminated at the first base because the sequence is read from bottom to top. The band that travelled the second-farthest through the gel must have ended at the base that came second in the sequence.
To make it easier to analyse the DNA sequence, ddNTPs are frequently dyed, i.e., labelled with a specific fluorescence. Modern technology can identify the multiple wavelengths at which dyed ddNTP fluoresces. A base-label diagram of the various wavelengths being analysed can thus be produced.
